[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
12














12
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
Github user jackylk commented on a diff in the pull request:
https://github.com/apache/carbondata/pull/2200#discussion_r183210908 --- Diff: datamap/bloom/src/main/java/org/apache/carbondata/datamap/bloom/BloomDataMapWriter.java --- @@ -0,0 +1,216 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.carbondata.datamap.bloom; + +import java.io.DataOutputStream; +import java.io.File; +import java.io.IOException; +import java.io.ObjectOutputStream; +import java.util.ArrayList; +import java.util.HashMap; +import java.util.List; +import java.util.Map; + +import org.apache.carbondata.common.annotations.InterfaceAudience; +import org.apache.carbondata.core.datamap.DataMapMeta; +import org.apache.carbondata.core.datamap.Segment; +import org.apache.carbondata.core.datamap.dev.DataMapWriter; +import org.apache.carbondata.core.datastore.impl.FileFactory; +import org.apache.carbondata.core.datastore.page.ColumnPage; +import org.apache.carbondata.core.metadata.AbsoluteTableIdentifier; +import org.apache.carbondata.core.metadata.datatype.DataType; +import org.apache.carbondata.core.metadata.datatype.DataTypes; +import org.apache.carbondata.core.util.CarbonUtil; + +import com.google.common.hash.BloomFilter; +import com.google.common.hash.Funnels; +import org.apache.hadoop.fs.FileSystem; +import org.apache.hadoop.fs.Path; + +@InterfaceAudience.Internal +public class BloomDataMapWriter extends DataMapWriter { + /** + * suppose one blocklet contains 20 page and all the indexed value is distinct. + * later we can make it configurable. + */ + private static final int BLOOM_FILTER_SIZE = 32000 * 20; + private String dataMapName; + private List<String> indexedColumns; + // map column name to ordinal in pages + private Map<String, Integer> col2Ordianl; + private Map<String, DataType> col2DataType; + private String currentBlockId; + private int currentBlockletId; + private List<String> currentDMFiles; + private List<DataOutputStream> currentDataOutStreams; + private List<ObjectOutputStream> currentObjectOutStreams; + private List<BloomFilter<byte[]>> indexBloomFilters; + + public BloomDataMapWriter(AbsoluteTableIdentifier identifier, DataMapMeta dataMapMeta, + Segment segment, String writeDirectoryPath) { + super(identifier, segment, writeDirectoryPath); + dataMapName = dataMapMeta.getDataMapName(); + indexedColumns = dataMapMeta.getIndexedColumns(); + col2Ordianl = new HashMap<String, Integer>(indexedColumns.size()); + col2DataType = new HashMap<String, DataType>(indexedColumns.size()); + + currentDMFiles = new ArrayList<String>(indexedColumns.size()); + currentDataOutStreams = new ArrayList<DataOutputStream>(indexedColumns.size()); + currentObjectOutStreams = new ArrayList<ObjectOutputStream>(indexedColumns.size()); + + indexBloomFilters = new ArrayList<BloomFilter<byte[]>>(indexedColumns.size()); + } + + @Override + public void onBlockStart(String blockId, long taskId) throws IOException { + this.currentBlockId = blockId; + this.currentBlockletId = 0; + currentDMFiles.clear(); + currentDataOutStreams.clear(); + currentObjectOutStreams.clear(); + initDataMapFile(); + } + + @Override + public void onBlockEnd(String blockId) throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + CarbonUtil.closeStreams(this.currentDataOutStreams.get(indexColId), + this.currentObjectOutStreams.get(indexColId)); + commitFile(this.currentDMFiles.get(indexColId)); + } + } + + @Override public void onBlockletStart(int blockletId) { + this.currentBlockletId = blockletId; + indexBloomFilters.clear(); + for (int i = 0; i < indexedColumns.size(); i++) { + indexBloomFilters.add(BloomFilter.create(Funnels.byteArrayFunnel(), + BLOOM_FILTER_SIZE, 0.00001d)); + } + } + + @Override + public void onBlockletEnd(int blockletId) { + try { + writeBloomDataMapFile(); + } catch (Exception e) { + for (ObjectOutputStream objectOutputStream : currentObjectOutStreams) { + CarbonUtil.closeStreams(objectOutputStream); + } + for (DataOutputStream dataOutputStream : currentDataOutStreams) { + CarbonUtil.closeStreams(dataOutputStream); + } + throw new RuntimeException(e); + } + } + + @Override public void onPageAdded(int blockletId, int pageId, ColumnPage[] pages) + throws IOException { + col2Ordianl.clear(); + col2DataType.clear(); + for (int colId = 0; colId < pages.length; colId++) { + String columnName = pages[colId].getColumnSpec().getFieldName().toLowerCase(); + if (indexedColumns.contains(columnName)) { --- End diff -- This is not needed. The input `pages` contains the indexed column only --- |
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
In reply to this post by qiuchenjian-2
Github user jackylk commented on a diff in the pull request:
https://github.com/apache/carbondata/pull/2200#discussion_r183210997 --- Diff: datamap/bloom/src/main/java/org/apache/carbondata/datamap/bloom/BloomDataMapWriter.java --- @@ -0,0 +1,216 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.carbondata.datamap.bloom; + +import java.io.DataOutputStream; +import java.io.File; +import java.io.IOException; +import java.io.ObjectOutputStream; +import java.util.ArrayList; +import java.util.HashMap; +import java.util.List; +import java.util.Map; + +import org.apache.carbondata.common.annotations.InterfaceAudience; +import org.apache.carbondata.core.datamap.DataMapMeta; +import org.apache.carbondata.core.datamap.Segment; +import org.apache.carbondata.core.datamap.dev.DataMapWriter; +import org.apache.carbondata.core.datastore.impl.FileFactory; +import org.apache.carbondata.core.datastore.page.ColumnPage; +import org.apache.carbondata.core.metadata.AbsoluteTableIdentifier; +import org.apache.carbondata.core.metadata.datatype.DataType; +import org.apache.carbondata.core.metadata.datatype.DataTypes; +import org.apache.carbondata.core.util.CarbonUtil; + +import com.google.common.hash.BloomFilter; +import com.google.common.hash.Funnels; +import org.apache.hadoop.fs.FileSystem; +import org.apache.hadoop.fs.Path; + +@InterfaceAudience.Internal +public class BloomDataMapWriter extends DataMapWriter { + /** + * suppose one blocklet contains 20 page and all the indexed value is distinct. + * later we can make it configurable. + */ + private static final int BLOOM_FILTER_SIZE = 32000 * 20; + private String dataMapName; + private List<String> indexedColumns; + // map column name to ordinal in pages + private Map<String, Integer> col2Ordianl; + private Map<String, DataType> col2DataType; + private String currentBlockId; + private int currentBlockletId; + private List<String> currentDMFiles; + private List<DataOutputStream> currentDataOutStreams; + private List<ObjectOutputStream> currentObjectOutStreams; + private List<BloomFilter<byte[]>> indexBloomFilters; + + public BloomDataMapWriter(AbsoluteTableIdentifier identifier, DataMapMeta dataMapMeta, + Segment segment, String writeDirectoryPath) { + super(identifier, segment, writeDirectoryPath); + dataMapName = dataMapMeta.getDataMapName(); + indexedColumns = dataMapMeta.getIndexedColumns(); + col2Ordianl = new HashMap<String, Integer>(indexedColumns.size()); + col2DataType = new HashMap<String, DataType>(indexedColumns.size()); + + currentDMFiles = new ArrayList<String>(indexedColumns.size()); + currentDataOutStreams = new ArrayList<DataOutputStream>(indexedColumns.size()); + currentObjectOutStreams = new ArrayList<ObjectOutputStream>(indexedColumns.size()); + + indexBloomFilters = new ArrayList<BloomFilter<byte[]>>(indexedColumns.size()); + } + + @Override + public void onBlockStart(String blockId, long taskId) throws IOException { + this.currentBlockId = blockId; + this.currentBlockletId = 0; + currentDMFiles.clear(); + currentDataOutStreams.clear(); + currentObjectOutStreams.clear(); + initDataMapFile(); + } + + @Override + public void onBlockEnd(String blockId) throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + CarbonUtil.closeStreams(this.currentDataOutStreams.get(indexColId), + this.currentObjectOutStreams.get(indexColId)); + commitFile(this.currentDMFiles.get(indexColId)); + } + } + + @Override public void onBlockletStart(int blockletId) { + this.currentBlockletId = blockletId; + indexBloomFilters.clear(); + for (int i = 0; i < indexedColumns.size(); i++) { + indexBloomFilters.add(BloomFilter.create(Funnels.byteArrayFunnel(), + BLOOM_FILTER_SIZE, 0.00001d)); + } + } + + @Override + public void onBlockletEnd(int blockletId) { + try { + writeBloomDataMapFile(); + } catch (Exception e) { + for (ObjectOutputStream objectOutputStream : currentObjectOutStreams) { + CarbonUtil.closeStreams(objectOutputStream); + } + for (DataOutputStream dataOutputStream : currentDataOutStreams) { + CarbonUtil.closeStreams(dataOutputStream); + } + throw new RuntimeException(e); + } + } + + @Override public void onPageAdded(int blockletId, int pageId, ColumnPage[] pages) + throws IOException { + col2Ordianl.clear(); + col2DataType.clear(); + for (int colId = 0; colId < pages.length; colId++) { + String columnName = pages[colId].getColumnSpec().getFieldName().toLowerCase(); + if (indexedColumns.contains(columnName)) { + col2Ordianl.put(columnName, colId); + DataType columnType = pages[colId].getColumnSpec().getSchemaDataType(); + col2DataType.put(columnName, columnType); + } + } + + // for each row + for (int rowId = 0; rowId < pages[0].getPageSize(); rowId++) { + // for each indexed column + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + String indexedCol = indexedColumns.get(indexColId); + byte[] indexValue; + if (DataTypes.STRING == col2DataType.get(indexedCol) + || DataTypes.BYTE_ARRAY == col2DataType.get(indexedCol)) { + byte[] originValue = (byte[]) pages[col2Ordianl.get(indexedCol)].getData(rowId); + indexValue = new byte[originValue.length - 2]; + System.arraycopy(originValue, 2, indexValue, 0, originValue.length - 2); + } else { + Object originValue = pages[col2Ordianl.get(indexedCol)].getData(rowId); + indexValue = CarbonUtil.getValueAsBytes(col2DataType.get(indexedCol), originValue); + } + + indexBloomFilters.get(indexColId).put(indexValue); + } + } + } + + private void initDataMapFile() throws IOException { + String dataMapDir = genDataMapStorePath(this.writeDirectoryPath, this.dataMapName); + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + String dmFile = dataMapDir + File.separator + this.currentBlockId + + '.' + indexedColumns.get(indexColId) + BloomCoarseGrainDataMap.BLOOM_INDEX_SUFFIX; + DataOutputStream dataOutStream = null; + ObjectOutputStream objectOutStream = null; + try { + FileFactory.createNewFile(dmFile, FileFactory.getFileType(dmFile)); + dataOutStream = FileFactory.getDataOutputStream(dmFile, + FileFactory.getFileType(dmFile)); + objectOutStream = new ObjectOutputStream(dataOutStream); + } catch (IOException e) { + CarbonUtil.closeStreams(objectOutStream, dataOutStream); + throw new IOException(e); + } + + this.currentDMFiles.add(dmFile); + this.currentDataOutStreams.add(dataOutStream); + this.currentObjectOutStreams.add(objectOutStream); + } + } + + private void writeBloomDataMapFile() throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + BloomDMModel model = new BloomDMModel(this.currentBlockId, this.currentBlockletId, + indexBloomFilters.get(indexColId)); + // only in higher version of guava-bloom-filter, it provides readFrom/writeTo interface. --- End diff -- why we can not use higher version of guava? which component prevents us from introduce higher version? which version? --- |
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
In reply to this post by qiuchenjian-2
Github user jackylk commented on a diff in the pull request:
https://github.com/apache/carbondata/pull/2200#discussion_r183211031 --- Diff: datamap/bloom/src/main/java/org/apache/carbondata/datamap/bloom/BloomDataMapWriter.java --- @@ -0,0 +1,216 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.carbondata.datamap.bloom; + +import java.io.DataOutputStream; +import java.io.File; +import java.io.IOException; +import java.io.ObjectOutputStream; +import java.util.ArrayList; +import java.util.HashMap; +import java.util.List; +import java.util.Map; + +import org.apache.carbondata.common.annotations.InterfaceAudience; +import org.apache.carbondata.core.datamap.DataMapMeta; +import org.apache.carbondata.core.datamap.Segment; +import org.apache.carbondata.core.datamap.dev.DataMapWriter; +import org.apache.carbondata.core.datastore.impl.FileFactory; +import org.apache.carbondata.core.datastore.page.ColumnPage; +import org.apache.carbondata.core.metadata.AbsoluteTableIdentifier; +import org.apache.carbondata.core.metadata.datatype.DataType; +import org.apache.carbondata.core.metadata.datatype.DataTypes; +import org.apache.carbondata.core.util.CarbonUtil; + +import com.google.common.hash.BloomFilter; +import com.google.common.hash.Funnels; +import org.apache.hadoop.fs.FileSystem; +import org.apache.hadoop.fs.Path; + +@InterfaceAudience.Internal +public class BloomDataMapWriter extends DataMapWriter { + /** + * suppose one blocklet contains 20 page and all the indexed value is distinct. + * later we can make it configurable. + */ + private static final int BLOOM_FILTER_SIZE = 32000 * 20; --- End diff -- Can you make one DMPROPERTY for it? Is it control the bloom filter size? --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user jackylk commented on the issue:
https://github.com/apache/carbondata/pull/2200 Thanks for working on this, it is a very good feature ð --- |
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
In reply to this post by qiuchenjian-2
Github user xuchuanyin commented on a diff in the pull request:
https://github.com/apache/carbondata/pull/2200#discussion_r183211712 --- Diff: datamap/bloom/src/main/java/org/apache/carbondata/datamap/bloom/BloomDataMapWriter.java --- @@ -0,0 +1,216 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.carbondata.datamap.bloom; + +import java.io.DataOutputStream; +import java.io.File; +import java.io.IOException; +import java.io.ObjectOutputStream; +import java.util.ArrayList; +import java.util.HashMap; +import java.util.List; +import java.util.Map; + +import org.apache.carbondata.common.annotations.InterfaceAudience; +import org.apache.carbondata.core.datamap.DataMapMeta; +import org.apache.carbondata.core.datamap.Segment; +import org.apache.carbondata.core.datamap.dev.DataMapWriter; +import org.apache.carbondata.core.datastore.impl.FileFactory; +import org.apache.carbondata.core.datastore.page.ColumnPage; +import org.apache.carbondata.core.metadata.AbsoluteTableIdentifier; +import org.apache.carbondata.core.metadata.datatype.DataType; +import org.apache.carbondata.core.metadata.datatype.DataTypes; +import org.apache.carbondata.core.util.CarbonUtil; + +import com.google.common.hash.BloomFilter; +import com.google.common.hash.Funnels; +import org.apache.hadoop.fs.FileSystem; +import org.apache.hadoop.fs.Path; + +@InterfaceAudience.Internal +public class BloomDataMapWriter extends DataMapWriter { + /** + * suppose one blocklet contains 20 page and all the indexed value is distinct. + * later we can make it configurable. + */ + private static final int BLOOM_FILTER_SIZE = 32000 * 20; + private String dataMapName; + private List<String> indexedColumns; + // map column name to ordinal in pages + private Map<String, Integer> col2Ordianl; + private Map<String, DataType> col2DataType; + private String currentBlockId; + private int currentBlockletId; + private List<String> currentDMFiles; + private List<DataOutputStream> currentDataOutStreams; + private List<ObjectOutputStream> currentObjectOutStreams; + private List<BloomFilter<byte[]>> indexBloomFilters; + + public BloomDataMapWriter(AbsoluteTableIdentifier identifier, DataMapMeta dataMapMeta, + Segment segment, String writeDirectoryPath) { + super(identifier, segment, writeDirectoryPath); + dataMapName = dataMapMeta.getDataMapName(); + indexedColumns = dataMapMeta.getIndexedColumns(); + col2Ordianl = new HashMap<String, Integer>(indexedColumns.size()); + col2DataType = new HashMap<String, DataType>(indexedColumns.size()); + + currentDMFiles = new ArrayList<String>(indexedColumns.size()); + currentDataOutStreams = new ArrayList<DataOutputStream>(indexedColumns.size()); + currentObjectOutStreams = new ArrayList<ObjectOutputStream>(indexedColumns.size()); + + indexBloomFilters = new ArrayList<BloomFilter<byte[]>>(indexedColumns.size()); + } + + @Override + public void onBlockStart(String blockId, long taskId) throws IOException { + this.currentBlockId = blockId; + this.currentBlockletId = 0; + currentDMFiles.clear(); + currentDataOutStreams.clear(); + currentObjectOutStreams.clear(); + initDataMapFile(); + } + + @Override + public void onBlockEnd(String blockId) throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + CarbonUtil.closeStreams(this.currentDataOutStreams.get(indexColId), + this.currentObjectOutStreams.get(indexColId)); + commitFile(this.currentDMFiles.get(indexColId)); + } + } + + @Override public void onBlockletStart(int blockletId) { + this.currentBlockletId = blockletId; + indexBloomFilters.clear(); + for (int i = 0; i < indexedColumns.size(); i++) { + indexBloomFilters.add(BloomFilter.create(Funnels.byteArrayFunnel(), + BLOOM_FILTER_SIZE, 0.00001d)); + } + } + + @Override + public void onBlockletEnd(int blockletId) { + try { + writeBloomDataMapFile(); + } catch (Exception e) { + for (ObjectOutputStream objectOutputStream : currentObjectOutStreams) { + CarbonUtil.closeStreams(objectOutputStream); + } + for (DataOutputStream dataOutputStream : currentDataOutStreams) { + CarbonUtil.closeStreams(dataOutputStream); + } + throw new RuntimeException(e); + } + } + + @Override public void onPageAdded(int blockletId, int pageId, ColumnPage[] pages) + throws IOException { + col2Ordianl.clear(); + col2DataType.clear(); + for (int colId = 0; colId < pages.length; colId++) { + String columnName = pages[colId].getColumnSpec().getFieldName().toLowerCase(); + if (indexedColumns.contains(columnName)) { + col2Ordianl.put(columnName, colId); + DataType columnType = pages[colId].getColumnSpec().getSchemaDataType(); + col2DataType.put(columnName, columnType); + } + } + + // for each row + for (int rowId = 0; rowId < pages[0].getPageSize(); rowId++) { + // for each indexed column + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + String indexedCol = indexedColumns.get(indexColId); + byte[] indexValue; + if (DataTypes.STRING == col2DataType.get(indexedCol) + || DataTypes.BYTE_ARRAY == col2DataType.get(indexedCol)) { + byte[] originValue = (byte[]) pages[col2Ordianl.get(indexedCol)].getData(rowId); + indexValue = new byte[originValue.length - 2]; + System.arraycopy(originValue, 2, indexValue, 0, originValue.length - 2); + } else { + Object originValue = pages[col2Ordianl.get(indexedCol)].getData(rowId); + indexValue = CarbonUtil.getValueAsBytes(col2DataType.get(indexedCol), originValue); + } + + indexBloomFilters.get(indexColId).put(indexValue); + } + } + } + + private void initDataMapFile() throws IOException { + String dataMapDir = genDataMapStorePath(this.writeDirectoryPath, this.dataMapName); + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + String dmFile = dataMapDir + File.separator + this.currentBlockId + + '.' + indexedColumns.get(indexColId) + BloomCoarseGrainDataMap.BLOOM_INDEX_SUFFIX; + DataOutputStream dataOutStream = null; + ObjectOutputStream objectOutStream = null; + try { + FileFactory.createNewFile(dmFile, FileFactory.getFileType(dmFile)); + dataOutStream = FileFactory.getDataOutputStream(dmFile, + FileFactory.getFileType(dmFile)); + objectOutStream = new ObjectOutputStream(dataOutStream); + } catch (IOException e) { + CarbonUtil.closeStreams(objectOutStream, dataOutStream); + throw new IOException(e); + } + + this.currentDMFiles.add(dmFile); + this.currentDataOutStreams.add(dataOutStream); + this.currentObjectOutStreams.add(objectOutStream); + } + } + + private void writeBloomDataMapFile() throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + BloomDMModel model = new BloomDMModel(this.currentBlockId, this.currentBlockletId, + indexBloomFilters.get(indexColId)); + // only in higher version of guava-bloom-filter, it provides readFrom/writeTo interface. --- End diff -- 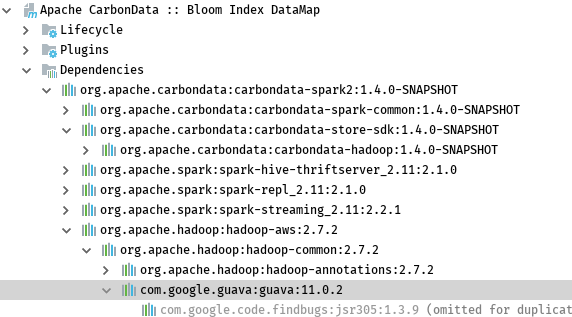 As shown above, hadoop-common use 11.0.2 version of guava. I've checked the interface of readFrom/writeTo of the bloomfilter provided by guava, it needs 24.* version. See https://github.com/google/guava/commit/62d17005a48e9b1044f1ed2d5de8905426d75299#diff-223d254389aa08fae7876742f8f07e8c for detailed information. Since hadoop-common rely on this package, So I not going to replace it. --- |
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
In reply to this post by qiuchenjian-2
Github user xuchuanyin commented on a diff in the pull request:
https://github.com/apache/carbondata/pull/2200#discussion_r183211728 --- Diff: datamap/bloom/src/main/java/org/apache/carbondata/datamap/bloom/BloomDataMapWriter.java --- @@ -0,0 +1,216 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.carbondata.datamap.bloom; + +import java.io.DataOutputStream; +import java.io.File; +import java.io.IOException; +import java.io.ObjectOutputStream; +import java.util.ArrayList; +import java.util.HashMap; +import java.util.List; +import java.util.Map; + +import org.apache.carbondata.common.annotations.InterfaceAudience; +import org.apache.carbondata.core.datamap.DataMapMeta; +import org.apache.carbondata.core.datamap.Segment; +import org.apache.carbondata.core.datamap.dev.DataMapWriter; +import org.apache.carbondata.core.datastore.impl.FileFactory; +import org.apache.carbondata.core.datastore.page.ColumnPage; +import org.apache.carbondata.core.metadata.AbsoluteTableIdentifier; +import org.apache.carbondata.core.metadata.datatype.DataType; +import org.apache.carbondata.core.metadata.datatype.DataTypes; +import org.apache.carbondata.core.util.CarbonUtil; + +import com.google.common.hash.BloomFilter; +import com.google.common.hash.Funnels; +import org.apache.hadoop.fs.FileSystem; +import org.apache.hadoop.fs.Path; + +@InterfaceAudience.Internal +public class BloomDataMapWriter extends DataMapWriter { + /** + * suppose one blocklet contains 20 page and all the indexed value is distinct. + * later we can make it configurable. + */ + private static final int BLOOM_FILTER_SIZE = 32000 * 20; --- End diff -- Yeah, it is used to control the rate. I'll make a default value for this. --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user CarbonDataQA commented on the issue:
https://github.com/apache/carbondata/pull/2200 Build Success with Spark 2.2.1, Please check CI http://88.99.58.216:8080/job/ApacheCarbonPRBuilder/4100/ --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user CarbonDataQA commented on the issue:
https://github.com/apache/carbondata/pull/2200 Build Failed with Spark 2.1.0, Please check CI http://136.243.101.176:8080/job/ApacheCarbonPRBuilder1/5280/ --- |
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
In reply to this post by qiuchenjian-2
Github user jackylk commented on a diff in the pull request:
https://github.com/apache/carbondata/pull/2200#discussion_r183212032 --- Diff: datamap/bloom/src/main/java/org/apache/carbondata/datamap/bloom/BloomDataMapWriter.java --- @@ -0,0 +1,216 @@ +/* + * Licensed to the Apache Software Foundation (ASF) under one or more + * contributor license agreements. See the NOTICE file distributed with + * this work for additional information regarding copyright ownership. + * The ASF licenses this file to You under the Apache License, Version 2.0 + * (the "License"); you may not use this file except in compliance with + * the License. You may obtain a copy of the License at + * + * http://www.apache.org/licenses/LICENSE-2.0 + * + * Unless required by applicable law or agreed to in writing, software + * distributed under the License is distributed on an "AS IS" BASIS, + * WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. + * See the License for the specific language governing permissions and + * limitations under the License. + */ +package org.apache.carbondata.datamap.bloom; + +import java.io.DataOutputStream; +import java.io.File; +import java.io.IOException; +import java.io.ObjectOutputStream; +import java.util.ArrayList; +import java.util.HashMap; +import java.util.List; +import java.util.Map; + +import org.apache.carbondata.common.annotations.InterfaceAudience; +import org.apache.carbondata.core.datamap.DataMapMeta; +import org.apache.carbondata.core.datamap.Segment; +import org.apache.carbondata.core.datamap.dev.DataMapWriter; +import org.apache.carbondata.core.datastore.impl.FileFactory; +import org.apache.carbondata.core.datastore.page.ColumnPage; +import org.apache.carbondata.core.metadata.AbsoluteTableIdentifier; +import org.apache.carbondata.core.metadata.datatype.DataType; +import org.apache.carbondata.core.metadata.datatype.DataTypes; +import org.apache.carbondata.core.util.CarbonUtil; + +import com.google.common.hash.BloomFilter; +import com.google.common.hash.Funnels; +import org.apache.hadoop.fs.FileSystem; +import org.apache.hadoop.fs.Path; + +@InterfaceAudience.Internal +public class BloomDataMapWriter extends DataMapWriter { + /** + * suppose one blocklet contains 20 page and all the indexed value is distinct. + * later we can make it configurable. + */ + private static final int BLOOM_FILTER_SIZE = 32000 * 20; + private String dataMapName; + private List<String> indexedColumns; + // map column name to ordinal in pages + private Map<String, Integer> col2Ordianl; + private Map<String, DataType> col2DataType; + private String currentBlockId; + private int currentBlockletId; + private List<String> currentDMFiles; + private List<DataOutputStream> currentDataOutStreams; + private List<ObjectOutputStream> currentObjectOutStreams; + private List<BloomFilter<byte[]>> indexBloomFilters; + + public BloomDataMapWriter(AbsoluteTableIdentifier identifier, DataMapMeta dataMapMeta, + Segment segment, String writeDirectoryPath) { + super(identifier, segment, writeDirectoryPath); + dataMapName = dataMapMeta.getDataMapName(); + indexedColumns = dataMapMeta.getIndexedColumns(); + col2Ordianl = new HashMap<String, Integer>(indexedColumns.size()); + col2DataType = new HashMap<String, DataType>(indexedColumns.size()); + + currentDMFiles = new ArrayList<String>(indexedColumns.size()); + currentDataOutStreams = new ArrayList<DataOutputStream>(indexedColumns.size()); + currentObjectOutStreams = new ArrayList<ObjectOutputStream>(indexedColumns.size()); + + indexBloomFilters = new ArrayList<BloomFilter<byte[]>>(indexedColumns.size()); + } + + @Override + public void onBlockStart(String blockId, long taskId) throws IOException { + this.currentBlockId = blockId; + this.currentBlockletId = 0; + currentDMFiles.clear(); + currentDataOutStreams.clear(); + currentObjectOutStreams.clear(); + initDataMapFile(); + } + + @Override + public void onBlockEnd(String blockId) throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + CarbonUtil.closeStreams(this.currentDataOutStreams.get(indexColId), + this.currentObjectOutStreams.get(indexColId)); + commitFile(this.currentDMFiles.get(indexColId)); + } + } + + @Override public void onBlockletStart(int blockletId) { + this.currentBlockletId = blockletId; + indexBloomFilters.clear(); + for (int i = 0; i < indexedColumns.size(); i++) { + indexBloomFilters.add(BloomFilter.create(Funnels.byteArrayFunnel(), + BLOOM_FILTER_SIZE, 0.00001d)); + } + } + + @Override + public void onBlockletEnd(int blockletId) { + try { + writeBloomDataMapFile(); + } catch (Exception e) { + for (ObjectOutputStream objectOutputStream : currentObjectOutStreams) { + CarbonUtil.closeStreams(objectOutputStream); + } + for (DataOutputStream dataOutputStream : currentDataOutStreams) { + CarbonUtil.closeStreams(dataOutputStream); + } + throw new RuntimeException(e); + } + } + + @Override public void onPageAdded(int blockletId, int pageId, ColumnPage[] pages) + throws IOException { + col2Ordianl.clear(); + col2DataType.clear(); + for (int colId = 0; colId < pages.length; colId++) { + String columnName = pages[colId].getColumnSpec().getFieldName().toLowerCase(); + if (indexedColumns.contains(columnName)) { + col2Ordianl.put(columnName, colId); + DataType columnType = pages[colId].getColumnSpec().getSchemaDataType(); + col2DataType.put(columnName, columnType); + } + } + + // for each row + for (int rowId = 0; rowId < pages[0].getPageSize(); rowId++) { + // for each indexed column + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + String indexedCol = indexedColumns.get(indexColId); + byte[] indexValue; + if (DataTypes.STRING == col2DataType.get(indexedCol) + || DataTypes.BYTE_ARRAY == col2DataType.get(indexedCol)) { + byte[] originValue = (byte[]) pages[col2Ordianl.get(indexedCol)].getData(rowId); + indexValue = new byte[originValue.length - 2]; + System.arraycopy(originValue, 2, indexValue, 0, originValue.length - 2); + } else { + Object originValue = pages[col2Ordianl.get(indexedCol)].getData(rowId); + indexValue = CarbonUtil.getValueAsBytes(col2DataType.get(indexedCol), originValue); + } + + indexBloomFilters.get(indexColId).put(indexValue); + } + } + } + + private void initDataMapFile() throws IOException { + String dataMapDir = genDataMapStorePath(this.writeDirectoryPath, this.dataMapName); + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + String dmFile = dataMapDir + File.separator + this.currentBlockId + + '.' + indexedColumns.get(indexColId) + BloomCoarseGrainDataMap.BLOOM_INDEX_SUFFIX; + DataOutputStream dataOutStream = null; + ObjectOutputStream objectOutStream = null; + try { + FileFactory.createNewFile(dmFile, FileFactory.getFileType(dmFile)); + dataOutStream = FileFactory.getDataOutputStream(dmFile, + FileFactory.getFileType(dmFile)); + objectOutStream = new ObjectOutputStream(dataOutStream); + } catch (IOException e) { + CarbonUtil.closeStreams(objectOutStream, dataOutStream); + throw new IOException(e); + } + + this.currentDMFiles.add(dmFile); + this.currentDataOutStreams.add(dataOutStream); + this.currentObjectOutStreams.add(objectOutStream); + } + } + + private void writeBloomDataMapFile() throws IOException { + for (int indexColId = 0; indexColId < indexedColumns.size(); indexColId++) { + BloomDMModel model = new BloomDMModel(this.currentBlockId, this.currentBlockletId, + indexBloomFilters.get(indexColId)); + // only in higher version of guava-bloom-filter, it provides readFrom/writeTo interface. --- End diff -- ok --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user xuchuanyin commented on the issue:
https://github.com/apache/carbondata/pull/2200 @jackylk review comments are fixed. Add a dm_property called âbloom_sizeâ to configure the size of bloom filter. Since we can have multiple indexed columns and the cardinality of them are differs, so we can support specifying corresponding bloom_size in the future. Besides, more tests are needed, such as exception, datamap profiling for this datamap -- will do it in the future, this version will provide the basic correct test case. --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user CarbonDataQA commented on the issue:
https://github.com/apache/carbondata/pull/2200 Build Success with Spark 2.2.1, Please check CI http://88.99.58.216:8080/job/ApacheCarbonPRBuilder/4101/ --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user CarbonDataQA commented on the issue:
https://github.com/apache/carbondata/pull/2200 Build Success with Spark 2.1.0, Please check CI http://136.243.101.176:8080/job/ApacheCarbonPRBuilder1/5281/ --- |
[GitHub] carbondata issue #2200: [CARBONDATA-2373][DataMap] Add bloom datamap to supp...
|
In reply to this post by qiuchenjian-2
Github user jackylk commented on the issue:
https://github.com/apache/carbondata/pull/2200 @xuchuanyin Agree. Need more enhancement in future. Thanks for working for this. LGTM --- |
[GitHub] carbondata pull request #2200: [CARBONDATA-2373][DataMap] Add bloom datamap ...
|
In reply to this post by qiuchenjian-2
|
«
Return to Apache CarbonData JIRA issues
|
1 view|%1 views
| Free forum by Nabble | Edit this page |

